-
PDF
- Split View
-
Views
-
Cite
Cite
Shanmugam Saravanan, Madhavan Vidya, Pachamuthu Balakrishnan, Rami Kantor, Sunil S. Solomon, David Katzenstein, Nagalingeswaran Kumarasamy, Tokugha Yeptomi, Sathasivam Sivamalar, Samara Rifkin, Kenneth H. Mayer, Suniti Solomon, Viremia and HIV-1 Drug Resistance Mutations Among Patients Receiving Second-Line Highly Active Antiretroviral Therapy in Chennai, Southern India, Clinical Infectious Diseases, Volume 54, Issue 7, 1 April 2012, Pages 995–1000, https://doi.org/10.1093/cid/cir967
Close - Share Icon Share
Abstract
Background. A cross-sectional study among individuals receiving second-line antiretroviral treatment was conducted to report on the level of detectable viremia and the types of drug resistance mutations among those with detectable human immunodeficiency virus (HIV) type 1 plasma viral loads (PVLs).
Methods. PVLs were measured using Abbott m2000rt real-time polymerase chain reaction, and genotyping was performed with the ViroSeq genotyping system, version 2.0, and ViroSeq analysis software, version 2.8.
Results. Of 107 patient plasma specimens consecutively analyzed, 30 (28%) had undetectable PVLs (<150 copies/mL), and 77 (72%) were viremic with a median PVL of 5450 copies/mL (interquartile range, 169–1 997 967). Sequencing was done for 107 samples with PVLs >2000 copies/mL: 33 patients (73%) had 1 of the protease (PR) inhibitor mutations; 41 (91%) had nucleoside reverse-transcriptase inhibitor (NRTI) mutations; 33 (73%) had non-NRTI (NNRTI) mutations; and 30 (66.7%) had both NRTI and NNRTI mutations. Triple-class resistance to NRTIs, NNRTIs, and PR inhibitors was observed in 24 (53%) patients. Based on the mutational profiles observed, all 45 sequences were susceptible to darunavir and tipranavir, whereas 47% showed resistance to lopinavir, 58% showed resistance to atazanavir, and >60% showed resistance to saquinavir, indinavir, nelfinavir, and fosamprenavir.
Conclusions. The results of the study showed that the majority of patients receiving second-line antiretroviral therapy started to accumulate PR resistance mutations, and the mutation profiles suggest that darunavir might be the drug of choice for third-line regimens in India.
The human immunodeficiency virus (HIV) type 1 protease (PR), essential for the viral replication, plays a crucial role in the HIV-1 life cycle, consequently serving as an important target for antiretroviral therapy (ART). To date, many PR inhibitors (PIs), namely indinavir (IDV), ritonavir (RTV), saquinavir (SQV), nelfinavir (NFV), fosamprenavir (FPV), lopinavir (LPV), atazanavir (ATV), and darunavir (DRV), have been approved by the US Food and Drug Administration and are commercially available. In India, these drugs are often used as single agents with 2 nucleoside reverse-transcriptase inhibitors (NRTIs) as second-line treatment for those in whom first-line non-NRTI (NNRTI)–based therapies fail. Since 2005, boosted PIs are recommended in combination with low-dose RTV, the latter serving to increase the level of the companion PI. When PI-based ART fails to be fully suppressive, viral variants to PIs can emerge [1–5].
Resistance to PIs is mediated by the sequential appearance of amino acid substitutions at positions either in direct contact with the inhibitor or at distant sites that reduces the binding affinity between the inhibitor and the mutant PR enzyme [6]. The amino acid substitutions, which are defined as major mutations, may deeply impair the PR catalytic activity and, consequently, the replication capacity of the virus [7, 8], whereas the presence of compensatory mutations restores the replication capacity [7, 9]. Furthermore, most drug resistance mutations affecting the PR confer cross-resistance to other PIs in the class and are considered to be class specific rather than drug specific [10, 11].
The introduction of generic first-line drugs and improved access among HIV-infected patients has substantially reduced the morbidity and mortality of the infection in India. However, in resource-limited settings, limited access to plasma viral load (PVL) monitoring remains a challenge [12, 13]. Owing to the lack of PVL monitoring, early virologic failure goes undetected and the patients continue to be on the failing regimen until they develop immunologic failure. In the time between virologic failure and immunologic failure, the virus accumulates resistance mutations [14]. High levels of drug resistance to first-line drugs have been reported from various parts of the country [4, 14–17], and, as a result, patients are switched to PI-based, second-line regimens.
Although India has seen increased access to second-line antiretrovirals since January 2009, there are few studies on PR drug resistance to these drugs from this region [4, 5, 17]. There are many studies from Western countries on the subtype B, but HIV-1 subtypes differ from one another and could influence the spectrum of mutations that develop during selective drug pressure. For instance, L90M is selected in excess, whereas there is underrepresentation of D30N in subtype C compared with subtype B, which might have consequences for therapeutic decision making [18]. Furthermore, subtype-associated natural polymorphisms have the potential factors to influence rate and mutational pathways leading to drug resistance [19].
The current study was done at YRG Centre for AIDS Research and Education (YRG CARE), Voluntary Health Services (VHS), Chennai, a tertiary HIV referral center for Southern India, and has provided a continuum of care for >12 000 individuals infected with HIV. All patients are treated according to World Health Organization (WHO) treatment guidelines and are advised to initiate highly active ART before CD4 cell counts drop to <200 cells/μL or when they range between 200 and 350 cells/μL with an AIDS-defining illness. Patients are seen every 3 months or as clinically indicated. Current treatment guidelines in India recommend the initiation of RTV-boosted, second-line PIs, such as ATV, SQV, LPV, and IDV, combined with 2 NRTIs after treatment failure of NNRTI-based first-line therapy [20], which is in accordance with current WHO and US Department of Health and Human Services recommendations. This study aims to characterize the pattern of PR resistance mutations among Southern Indian patients who have been exposed to PI-based, second-line therapy.
SUBJECTS AND METHODS
A cross-sectional study of 107 patients infected with HIV-1, in whom first-line treatment had failed and who had received second-line boosted PIs for >6 months, were consecutively enrolled at YRG CARE between 2008 and 2009. All of the plasma specimens collected were screened for their viral load by Abbott m2000rt real-time polymerase chain reaction (Abbott Molecular Diagnostics), using the 0.2 uL manual extraction protocol with a detection limit of 150 to 10 million copies/mL. HIV-1 genotyping was performed in patients with a detectable PVL (>2000 copies/mL), using the ViroSeq genotyping system (version 2.0) with analysis software (version 2.8). The treatment history for all study patients was obtained from the YRG CARE natural history database. Reference sequences were obtained from Los Alamos National Laboratory online database (http://www.hiv.lanl.gov). Stanford HIV Drug Resistance online software, version 6.0.9 (http://hivdb.stanford.edu), was used to interpret the resistance mutations. Nucleic acid sequences were aligned pairwise using Clustal W in Molecular Evolutionary Genetic Analysis (MEGA) Software for Windows, version 4.0 [21].
The χ2 test and Fisher’s exact test were used to compare categorical variables, as appropriate. All analyses were performed using SPSS version 13.0 (SPSS). All tests of statistical significance were 2 sided, and differences were considered significant at P < .05.
RESULTS
During this study, all of the patients were receiving an RTV-boosted PI regimen. Most of them started with an ATV-containing PI regimen (47.6%), followed by an IDV-containing PI regimen (44.8%) and an LPV-containing PI regimen (7.5%). The major NRTI backbone consisted of a tenofovir-emtricitabine regimen (57%) (Table 1). The most common first-line regimens included AZT-3TC-NVP (38%), followed by d4t-3TC-NVP (33%), and AZT-3TC-EFV (8%), and the median time until switching of regimens was 25 months (interquartile range, 13–50 months). The treatment history and demographic details are given in Table 1.
Demographics and Treatment History of Patients Classified as ”Undetectable PVL” or Viremic
| Variable | Patients With Undetectable PVL (n = 30) | Viremic Patients (n = 77) |
| Sex, No. | ||
| Male | 24 | 63 |
| Female | 6 | 14 |
| Type of boosted PI, No. | ||
| ATV based | 14 | 37 |
| IDV based | 13 | 35 |
| LPV based | 3 | 5 |
| NRTI backbone, No. | ||
| TDF-FTC based | 18 | 43 |
| AZT-ddI | 3 | 12 |
| 3TC-ddI | 7 | 18 |
| AZT-3TC | 2 | 4 |
| History of unboosted PI, No. | 7 | 9 |
| NFV | 5 | 6 |
| IDV | 2 | 3 |
| CD4 cell count, median (IQR), cells/μL | ||
| Baseline | 92 (11–427) | 154 (11–888) |
| During switch to PI | 157 (14–448) | 100 (6–720) |
| At genotyping | 402 (14–938) | 190 (8–731) |
| Plasma viral load at genotyping, median (IQR), copies/mL | ||
| Undetectable levels (<150 copies/mL) | 30 | NA |
| 150 to <2000 copies/mL | NA | 627 (169–1987); n = 32 |
| >2000 to <5000 copies/mL | NA | 2562 (2188–2948); n = 5 |
| >5000 copies/mL | NA | 65 363 (5278–1 997 967); n = 40 |
| Variable | Patients With Undetectable PVL (n = 30) | Viremic Patients (n = 77) |
| Sex, No. | ||
| Male | 24 | 63 |
| Female | 6 | 14 |
| Type of boosted PI, No. | ||
| ATV based | 14 | 37 |
| IDV based | 13 | 35 |
| LPV based | 3 | 5 |
| NRTI backbone, No. | ||
| TDF-FTC based | 18 | 43 |
| AZT-ddI | 3 | 12 |
| 3TC-ddI | 7 | 18 |
| AZT-3TC | 2 | 4 |
| History of unboosted PI, No. | 7 | 9 |
| NFV | 5 | 6 |
| IDV | 2 | 3 |
| CD4 cell count, median (IQR), cells/μL | ||
| Baseline | 92 (11–427) | 154 (11–888) |
| During switch to PI | 157 (14–448) | 100 (6–720) |
| At genotyping | 402 (14–938) | 190 (8–731) |
| Plasma viral load at genotyping, median (IQR), copies/mL | ||
| Undetectable levels (<150 copies/mL) | 30 | NA |
| 150 to <2000 copies/mL | NA | 627 (169–1987); n = 32 |
| >2000 to <5000 copies/mL | NA | 2562 (2188–2948); n = 5 |
| >5000 copies/mL | NA | 65 363 (5278–1 997 967); n = 40 |
Abbreviations: 3TC, lamivudine; ATV, atazanavir; AZT, zidovudine; ddI, didanosine; FTC, emtricitabine; IDV, indinavir; IQR, interquartile range; LPV, lopinavir; NA, not applicable; NFV, nelfinavir; NRTI, nucleoside reverse-transcriptase inhibitor; PI, protease inhibitor; PVL, plasma viral load; TDF, tenofovir.
Demographics and Treatment History of Patients Classified as ”Undetectable PVL” or Viremic
| Variable | Patients With Undetectable PVL (n = 30) | Viremic Patients (n = 77) |
| Sex, No. | ||
| Male | 24 | 63 |
| Female | 6 | 14 |
| Type of boosted PI, No. | ||
| ATV based | 14 | 37 |
| IDV based | 13 | 35 |
| LPV based | 3 | 5 |
| NRTI backbone, No. | ||
| TDF-FTC based | 18 | 43 |
| AZT-ddI | 3 | 12 |
| 3TC-ddI | 7 | 18 |
| AZT-3TC | 2 | 4 |
| History of unboosted PI, No. | 7 | 9 |
| NFV | 5 | 6 |
| IDV | 2 | 3 |
| CD4 cell count, median (IQR), cells/μL | ||
| Baseline | 92 (11–427) | 154 (11–888) |
| During switch to PI | 157 (14–448) | 100 (6–720) |
| At genotyping | 402 (14–938) | 190 (8–731) |
| Plasma viral load at genotyping, median (IQR), copies/mL | ||
| Undetectable levels (<150 copies/mL) | 30 | NA |
| 150 to <2000 copies/mL | NA | 627 (169–1987); n = 32 |
| >2000 to <5000 copies/mL | NA | 2562 (2188–2948); n = 5 |
| >5000 copies/mL | NA | 65 363 (5278–1 997 967); n = 40 |
| Variable | Patients With Undetectable PVL (n = 30) | Viremic Patients (n = 77) |
| Sex, No. | ||
| Male | 24 | 63 |
| Female | 6 | 14 |
| Type of boosted PI, No. | ||
| ATV based | 14 | 37 |
| IDV based | 13 | 35 |
| LPV based | 3 | 5 |
| NRTI backbone, No. | ||
| TDF-FTC based | 18 | 43 |
| AZT-ddI | 3 | 12 |
| 3TC-ddI | 7 | 18 |
| AZT-3TC | 2 | 4 |
| History of unboosted PI, No. | 7 | 9 |
| NFV | 5 | 6 |
| IDV | 2 | 3 |
| CD4 cell count, median (IQR), cells/μL | ||
| Baseline | 92 (11–427) | 154 (11–888) |
| During switch to PI | 157 (14–448) | 100 (6–720) |
| At genotyping | 402 (14–938) | 190 (8–731) |
| Plasma viral load at genotyping, median (IQR), copies/mL | ||
| Undetectable levels (<150 copies/mL) | 30 | NA |
| 150 to <2000 copies/mL | NA | 627 (169–1987); n = 32 |
| >2000 to <5000 copies/mL | NA | 2562 (2188–2948); n = 5 |
| >5000 copies/mL | NA | 65 363 (5278–1 997 967); n = 40 |
Abbreviations: 3TC, lamivudine; ATV, atazanavir; AZT, zidovudine; ddI, didanosine; FTC, emtricitabine; IDV, indinavir; IQR, interquartile range; LPV, lopinavir; NA, not applicable; NFV, nelfinavir; NRTI, nucleoside reverse-transcriptase inhibitor; PI, protease inhibitor; PVL, plasma viral load; TDF, tenofovir.
The majority of the sequences were monophyletically clustered with subtype C (44 of 45; 98%), and 1 was subtype A (2%). Of the 107 patients, 30 (28%) had undetectable PVLs (<150 copies/mL), and 77 (72%) were viremic, with a median PVL of 5450 copies/mL (interquartile range, 169–1 997 967). Among the 77 viremic patients, sequencing was done for 45 samples with PVLs >2000 copies/mL (Table 1); the predominant PR mutations were M46I (22 samples; 49%), I54 V/A (17; 38%), V82A (17; 38%), and L90M (14; 31%). Among the reverse-transcriptase mutations, 40 (89%) had M184V, the predominant mutation, followed by 32 (71%) with thymidine-analog mutations (TAMs), of which 16 (50%) developed TAM1 (T215Y, L210W, and M41L), 9 (28%) developed TAM 2 (T215F, K70R, K219E, and D67N), and 7 (22%) had a mix of both TAM1 and 2 pathways. NNRTI mutations were observed in 33 (73%) patients, and 24 (53%) had triple-class resistance mutations. When etravirine (ETR) susceptibility was analyzed among these patients (using International AIDS Society 2010 guidelines [22]), the majority of them (32 of 45; 71%) were predicted to respond to ETR, whereas an intermediate to high level of resistance was observed in 29% (13 of 45).
Twelve patients (26.6%) had any one of the DRV resistance-associated mutations: G73S was observed in 8 patients (17.7%), I84V in 2 patients (4.4%), and L33F, L76V, and L89V in 1 patient (2.2%) each. None of the patients had >3 combinations of DRV-associated mutations, which confer resistance to DRV.
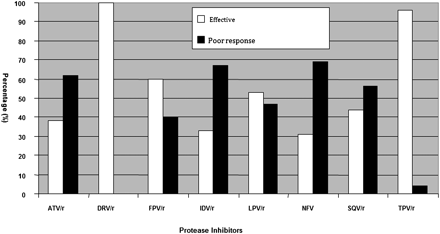
A higher rate of major PI resistance mutations was observed among patients receiving boosted IDV (IDV/r) than among those receiving boosted ATV (ATV/r), with rates of V82A (55% and 30%), L90M (35% and 26%), and I54V (45% and 35%) higher in the former group (P < .05). Intermediate resistance to LPV was observed in 9 of 23 patients (39%) receiving ATV/r and in 12 of 20 (60%) receiving IDV/r. The LPV resistance mutation patterns observed in ATV/r- and IDV/r-exposed groups were M46I + V82A/F + I54V (5/23 [22%] and 9/20 [45%]) and M46I/I54V + V82A (4/23 [17%] and 3/20 [15%]), respectively. Resistance was lowest to DRV (0%), followed by TPV (2%) and LPV (47%) (Figure 1). In the 45 patients genotyped, CD4 cell counts were <200 cells/μL in 34 and >200 cells/μL in 11. There was no significant difference (P = .1) between these groups in the pattern of mutations observed.
Summary and interpretation of genotypic resistance to protease inhibitors. A cumulative mutation score of 1–29 is considered effective (white), whereas scores >30 are taken as poor response to the drug (black), according to the online Stanford HIV database. Abbreviations: ATV, atazanavir; DRV, darunavir; FPV, fosamprenavir; IDV, indinavir; LPV, lopinavir; NFV, nelfinavir; SQV, saquinavir; TPV, tipranavir.
DISCUSSION
The current study analyzed the pattern of PR resistance mutations among HIV-1-infected patients exposed to second-line PR-based drugs. The PVL for the majority (63%) of the patients was either suppressed or below 5000 copies/mL, which is the threshold for virologic failure according to the WHO [23]. Among the 45 patients who were genotyped, 13 of the 32 mutations have been shown to reduce susceptibility to PIs as per the stanford resistance database, was observed.
A majority of the patients harbored TAMs, and TAM1 was the predominant pathway selected (>50%). This is because all of the patients had been exposed to thymidine-analog drugs in the first-line regimen and had been exposed for a longer period because of the lack of virologic monitoring. Approximately half of the patients had begun a TDF-based second-line regimen and one-third were receiving a didanosine-based regimen, neither of which select for TAMs; few patients received a didanosine-AZT dose, which is rarely used in the developed world and raises concerns of developing TAMs. Detection of early treatment failure based on virologic monitoring would ensure that patients avoid severe clinical complications and would prevent unnecessary early switching.
Although NNRTIs had been discontinued in all of the study patients, 73% of the patients still harbored NNRTI mutations, similar to findings of another study [24] that reports the persistence of NNRTI mutations for >12 months after discontinuation of the drug. The persistence of these mutations could be due to the limited impact of NNRTI resistance mutations on viral fitness. K103N, Y181C, and G190A were the predominant NNRTI mutations observed, because these have little or no effect on replication capacity [25].
The present study reports a higher frequency of PR resistance mutations (73%) compared with findings in studies from India [4, 5, 17]. Possible reasons for a lower prevalence in the other studies include small sample size (<5 patients exposed to PIs) and <12 months of PI exposure. M46I was the predominant mutation, in agreement with other findings from India [4, 5, 17], and L90M was observed in the majority of the patients compared with D30N, consistent with the earlier studies on subtype C [18, 26, 27]. In the ATV/r-exposed group, the signature mutations, I50L and N88S, were observed in only 2 patients, whereas the majority of resistance to ATV was due to the selection of cross-resistance conferring mutations. Among the IDV/r-exposed group, V82A, I54V, and M46I signature mutations were predominantly selected as reported elsewhere [1, 28].
Although DRV-associated mutations were observed in 12 patients in the study, the majority of them (8 of 12) were only accessory mutations. According to POWER studies [29], ≥3 DRV-associated resistance mutations (V32I, I47V/A, I50V, I54M/L, L76V, I84V/A/C) presented with reduced susceptibility to the drug, but none of the patients in the present study had >3 DRV resistance mutations, indicating higher levels of susceptibility to this drug. This was true for patients with CD4 cell counts above or below 200 cells/μL, because no significant difference in PI mutation patterns was observed between these groups.
Among the secondary PR mutations, amino acid variants at 7 polymorphic positions (codons 10, 20, 36, 63, 71, 77, and 93) also make major contributions to drug resistance. Whereas these mutations do not cause drug resistance by themselves, some of them increase drug resistance when present together with other PR mutations. Some also compensate for the decrease in catalytic efficiency caused by other PR mutations [30]. L10I and A71L were predominantly observed in our study, similar to findings in other studies [4, 5, 31]. M36I/L and I93L mutations, which have been associated with resistance to PIs in subtype B, were present in samples from all patients, indicating that these are polymorphisms specifically associated with subtype C HIV-1 and are not related to PI susceptibility or resistance in this subtype [18].
The patterns of primary and secondary mutations selected by the use of PIs in different combinations and in successive regimens may affect future therapeutic options by reducing the genetic barrier to clinical resistance. The current pattern of PI mutations suggests that a majority of the patients remain susceptible to DRV/r, followed by TPV/r. Of the 2 pathways that contribute to LPV resistance [32, 33], the IDV-like pathway caused by mutations at positions M46I/L, I54V/T/A/S, and V82A/T/F/S [33, 34] was frequently observed among the ATV- and IDV-exposed groups. This could be because patients in this study had been increasingly substituted from IDV- to ATV-based regimens. Moreover, IDV-based PI regimens were the first available in India, and generic-boosted ATV- and LPV-based regimens were later made available.
The results of this study also showed that 24 of the 45 patients genotyped (53%) have already developed triple-class resistance, necessitating an urgent need by the national programs to develop policies for third-line therapies, which include new drugs likely to have anti-HIV activity, such as integrase inhibitors and second-generation NNRTI and PIs. Randomized clinical trials have been conducted in developed countries, and the results show that the combination of raltegravir, ETR, and DRV/r was well tolerated and was associated with the rate of virologic suppression similar to that expected in treatment-naive patient [35]. Although the availability of these drugs in these settings now and in the near future is uncertain, similar randomized clinical trials in these settings are a priority, because patients have already started to accumulate resistance to second-line drugs, and a step towards the development of generic drugs for the third line might overcome the cost involved.
The higher frequency of resistance mutations observed also raises concern about transmitted resistance. So far, studies from India have reported transmitted resistance to reverse-transcriptase drugs ranging from 0.0% to 78% and to PI drugs ranging from 2% to 20% [15, 36–43]. Good adherence and proper patient monitoring are important tools for better treatment outcomes.
The results of this study have limitations, and it might not be possible to draw conclusions that may affect India’s HIV program policy on the use of third-line therapies. Because of the small sample size and because most of the patients attending YRG CARE come from the southern part of India, it might not be appropriate to generalize from these observations.
At present, with few third-line antiretroviral options for subsequent therapy in India, DRV might be the choice for third-line regimens. In contrast, it is important to strengthen the development of technologies for affordable PVL monitoring for early detection of virologic failure, and genotype-guided switching regimens could improve treatment outcomes.
Notes
Acknowledgments.
We are most grateful to the clinical and laboratory staff at YRG CARE and VHS (Chennai, India) for their facilitation of the study.
Financial support.
This work was supported by a research grant from YRG CARE (Chennai, India).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.





Comments